Developing a greater understanding of how our gut microbiome transmits between people to maintain health
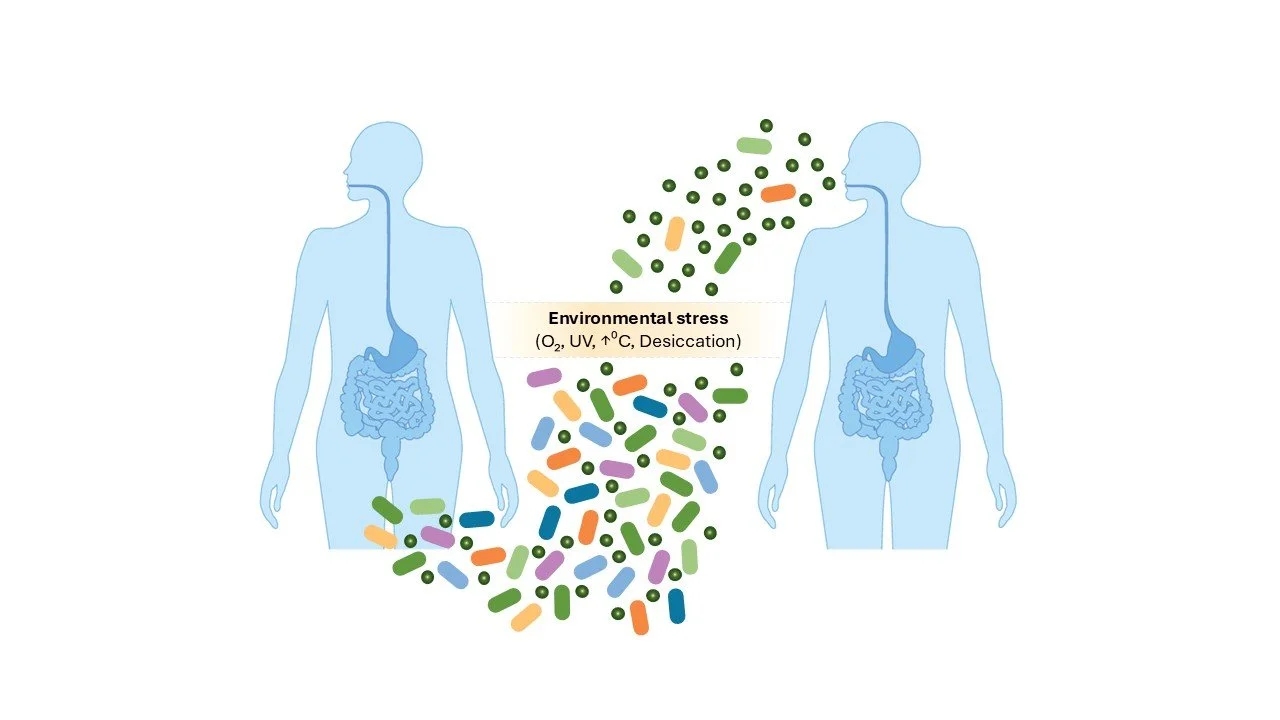
Our gut microbiome consists of diverse, beneficial microbial communities. All gut bacteria must transmit between and colonise new human hosts to survive. This also ensures we maintain the benefits of these bacteria including competing with harmful pathogens, metabolising carbohydrates indigestible to us and maintaining a balance in our immune system. But there’s a catch. Most human gut bacteria are strict anaerobes and quickly die in the presence of oxygen. However, we know transmission of gut bacteria between humans is constantly happening- from mother to baby during birth or shortly afterwards, between co-habiting family members or households and even between larger social networks. Our research seeks to understand how gut bacteria safely transmit and how this affects gut microbiome functions and evolution.
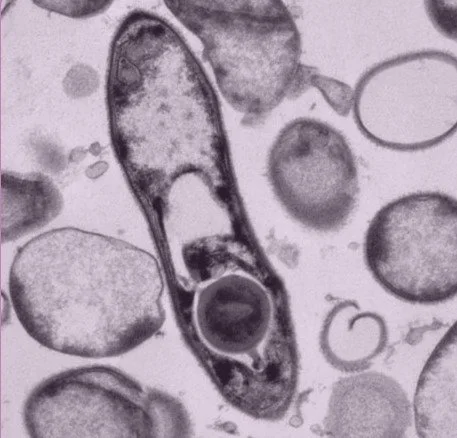
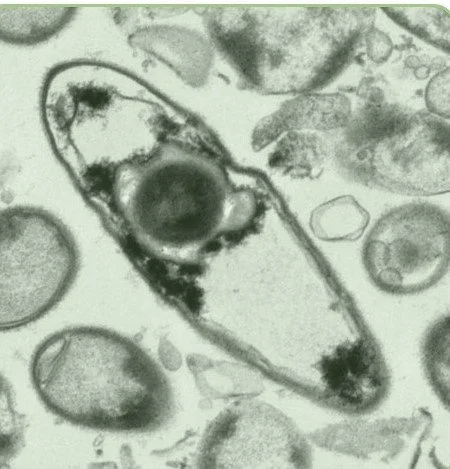
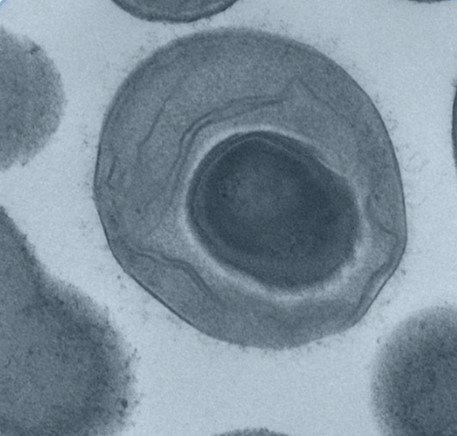
We focus on bacterial spores. These are produced by some Bacillota bacteria and are dormant and highly resilient structures that preserve bacterial DNA. Spores can tolerate oxygen, disinfectants and the temperature changes encountered outside of a human host for long periods of time. Once ingested by a new house, they recognise intestinal signals and spring back to life, re-forming a new bacterial cell that colonise the gut.
We use deeply-phenotyped culture collections of whole-genome sequenced gut bacteria as a platform for fundamental research and translational science and combine anaerobic microbiology and genomic approaches for detailed characterisation. We are interested in exploring the extent of sporulation in the gut microbiome, how this phenotype evolved, the genes involved and the triggers that initiate sporulation and germination. We also characterise spore-forming bacteria that can kill harmful pathogens.
In addition to sporulation, the lab also researches gut fungi and bacterial-fungal interactions. We are interested in understanding how bacteria and fungi compete and collaborate in the gut and how this knowledge can be used to identify commensal bacteria that can inhibit fungal pathogens.
Research in our lab is directly funded by the European Research Council (ERC Starter Grant) and Joint Programming Initiative on Antimicrobial Resistance (JPIAMR). If you are interested in joining us, please use Contact page for more information.
Main Research areas:
Gut microbiome sporulation processes
Evolution of sporulation and functional adaptations
Gut microbiome transmission
Fungal-bacterial interactions and inhibition of fungal pathogens
TEM image of bacterial vegetative cell with developing spore inside the mother cell. All TEM images created by Dave Goulding, Wellcome Sanger Institute.